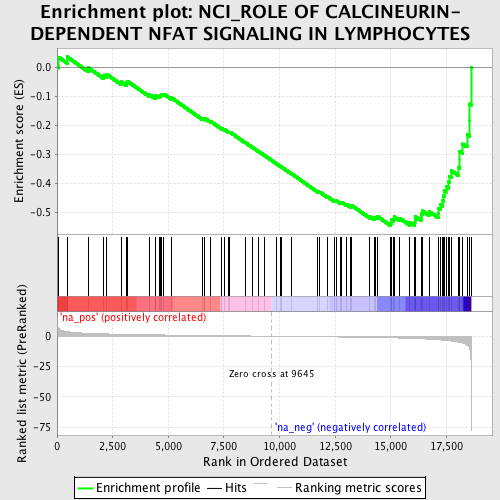
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | NCI_ROLE OF CALCINEURIN-DEPENDENT NFAT SIGNALING IN LYMPHOCYTES |
| Enrichment Score (ES) | -0.5468667 |
| Normalized Enrichment Score (NES) | -1.8486493 |
| Nominal p-value | 0.0013586957 |
| FDR q-value | 0.10135598 |
| FWER p-Value | 0.784 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IL5 | 54 | 6.551 | 0.0350 | No | ||
| 2 | CDK4 | 450 | 3.776 | 0.0355 | No | ||
| 3 | SLC3A2 | 1409 | 2.354 | -0.0025 | No | ||
| 4 | YWHAQ | 2088 | 1.942 | -0.0279 | No | ||
| 5 | NFATC2 | 2224 | 1.873 | -0.0243 | No | ||
| 6 | EGR3 | 2875 | 1.586 | -0.0502 | No | ||
| 7 | TBX21 | 3102 | 1.505 | -0.0537 | No | ||
| 8 | YWHAZ | 3177 | 1.476 | -0.0492 | No | ||
| 9 | PRKCD | 4150 | 1.173 | -0.0948 | No | ||
| 10 | BAD | 4407 | 1.108 | -0.1022 | No | ||
| 11 | RAN | 4431 | 1.102 | -0.0971 | No | ||
| 12 | TNF | 4598 | 1.062 | -0.0999 | No | ||
| 13 | E2F1 | 4642 | 1.052 | -0.0961 | No | ||
| 14 | RNF128 | 4690 | 1.039 | -0.0926 | No | ||
| 15 | PRKACA | 4799 | 1.014 | -0.0926 | No | ||
| 16 | IL4 | 5143 | 0.933 | -0.1057 | No | ||
| 17 | KPNB1 | 6534 | 0.622 | -0.1771 | No | ||
| 18 | YWHAB | 6626 | 0.604 | -0.1785 | No | ||
| 19 | XPO1 | 6628 | 0.603 | -0.1751 | No | ||
| 20 | PTGS2 | 6894 | 0.550 | -0.1862 | No | ||
| 21 | PRKCA | 7407 | 0.444 | -0.2112 | No | ||
| 22 | CASP3 | 7517 | 0.422 | -0.2147 | No | ||
| 23 | IL2RA | 7719 | 0.384 | -0.2233 | No | ||
| 24 | CAMK4 | 7761 | 0.376 | -0.2233 | No | ||
| 25 | PRKCZ | 8446 | 0.236 | -0.2589 | No | ||
| 26 | CALM1 | 8771 | 0.170 | -0.2754 | No | ||
| 27 | FKBP8 | 9068 | 0.113 | -0.2907 | No | ||
| 28 | CD40LG | 9328 | 0.064 | -0.3043 | No | ||
| 29 | YWHAE | 9858 | -0.041 | -0.3326 | No | ||
| 30 | YWHAG | 10062 | -0.083 | -0.3430 | No | ||
| 31 | KPNA2 | 10102 | -0.091 | -0.3446 | No | ||
| 32 | EGR4 | 10529 | -0.173 | -0.3666 | No | ||
| 33 | MAPK3 | 11722 | -0.435 | -0.4284 | No | ||
| 34 | FOS | 11810 | -0.455 | -0.4304 | No | ||
| 35 | PRKCE | 12144 | -0.521 | -0.4454 | No | ||
| 36 | FOSL1 | 12479 | -0.600 | -0.4599 | No | ||
| 37 | CSNK2A1 | 12552 | -0.617 | -0.4602 | No | ||
| 38 | CSF2 | 12718 | -0.657 | -0.4653 | No | ||
| 39 | NR4A1 | 12782 | -0.668 | -0.4649 | No | ||
| 40 | IL2 | 13006 | -0.720 | -0.4727 | No | ||
| 41 | CREBBP | 13192 | -0.773 | -0.4783 | No | ||
| 42 | IL3 | 13241 | -0.787 | -0.4763 | No | ||
| 43 | CALM2 | 14059 | -1.001 | -0.5146 | No | ||
| 44 | JUN | 14245 | -1.057 | -0.5184 | No | ||
| 45 | CBLB | 14325 | -1.083 | -0.5164 | No | ||
| 46 | PTPRK | 14407 | -1.106 | -0.5144 | No | ||
| 47 | MEF2D | 15003 | -1.317 | -0.5389 | Yes | ||
| 48 | PTPN1 | 15017 | -1.323 | -0.5319 | Yes | ||
| 49 | MAPK8 | 15028 | -1.330 | -0.5248 | Yes | ||
| 50 | GATA3 | 15140 | -1.374 | -0.5228 | Yes | ||
| 51 | IFNG | 15149 | -1.377 | -0.5153 | Yes | ||
| 52 | MAPK14 | 15392 | -1.490 | -0.5197 | Yes | ||
| 53 | POU2F1 | 15845 | -1.743 | -0.5340 | Yes | ||
| 54 | IRF4 | 16084 | -1.898 | -0.5359 | Yes | ||
| 55 | CSNK1A1 | 16088 | -1.901 | -0.5251 | Yes | ||
| 56 | MAP3K1 | 16099 | -1.908 | -0.5146 | Yes | ||
| 57 | PPARG | 16361 | -2.107 | -0.5165 | Yes | ||
| 58 | SFN | 16365 | -2.109 | -0.5044 | Yes | ||
| 59 | PRKCH | 16424 | -2.159 | -0.4951 | Yes | ||
| 60 | CTLA4 | 16721 | -2.425 | -0.4970 | Yes | ||
| 61 | MAPK9 | 17129 | -2.896 | -0.5022 | Yes | ||
| 62 | GSK3B | 17139 | -2.906 | -0.4859 | Yes | ||
| 63 | CALM3 | 17223 | -3.039 | -0.4728 | Yes | ||
| 64 | JUNB | 17330 | -3.198 | -0.4600 | Yes | ||
| 65 | PRKCQ | 17373 | -3.270 | -0.4434 | Yes | ||
| 66 | YWHAH | 17395 | -3.317 | -0.4253 | Yes | ||
| 67 | FKBP1A | 17483 | -3.475 | -0.4099 | Yes | ||
| 68 | ITCH | 17608 | -3.701 | -0.3952 | Yes | ||
| 69 | MAP3K8 | 17644 | -3.789 | -0.3752 | Yes | ||
| 70 | FASLG | 17717 | -3.934 | -0.3563 | Yes | ||
| 71 | NFATC1 | 18056 | -4.862 | -0.3464 | Yes | ||
| 72 | PIM1 | 18097 | -5.045 | -0.3194 | Yes | ||
| 73 | NFATC3 | 18098 | -5.049 | -0.2902 | Yes | ||
| 74 | FOXP3 | 18238 | -5.724 | -0.2646 | Yes | ||
| 75 | EGR1 | 18429 | -7.374 | -0.2322 | Yes | ||
| 76 | BCL2 | 18530 | -9.542 | -0.1824 | Yes | ||
| 77 | DGKA | 18533 | -9.632 | -0.1268 | Yes | ||
| 78 | EGR2 | 18610 | -22.681 | 0.0003 | Yes |
